Prof. Dr. Andreas Neueder


The team
Nathalie Birth (Technical assistant)
The research
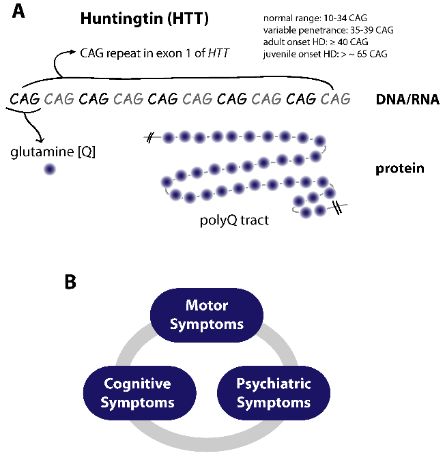
(A) The CAG repeat expansion mutation in HTT leads to a polyQ tract in the HTT protein. (B) HD manifests with a triade of motor (chorea), cognitive and psychiatric symptoms.
The expanded polyQ tract of mutated HTT results in toxic gains-of-function of the protein, the generation of small fragments of HTT, the appearance of aggregates and disruption of various cellular processes. Eventually these events lead to cell death, most prominent in the neurons of the striatum.
Read more here
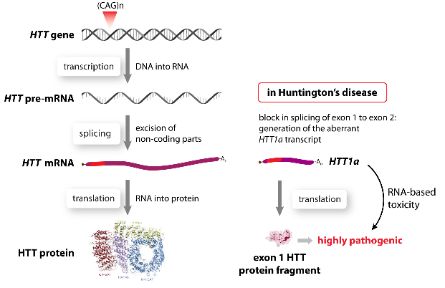
We have previously identified a novel mechanism that leads to the generation of the most toxic fragment of HTT: exon 1 HTT. This fragment consists only of exon 1 of the HTT gene including the polyQ tract. The generation of this fragment is based on a block in the correct splicing reaction of exon 1 HTT to exon 2 HTT resulting in the generation of a novel small RNA HTT1a. This RNA based pathogenic process occurs in all knock-in mouse models of HD, and most importantly also in human HD patients.
Read the publications here
contact
lab NEWS




Come and visit us here or contact us if want further information.


In the present study, we demonstrate that examining extracellular vesicles (EVs) can give new insights into pathologic mechanisms in Huntington disease.EVs from HD individuals convey specific actionable information in comparison to EVs from healthy people highlighting the biological relevance and potential use of EVs as a biomarker in clinical trials.
https://doi.org/10.1002/ctm2.1525

We show that life-long expression of mutant HTT causes a mitochondrial phenotype indicative of mtDNA instability in fresh post-mitotic human skeletal muscle. Thus, genomic instability may not be limited to nuclear DNA.
https://doi.org/10.1093/brain/awae007

Astrocytes are dysfunctional in HD patients and the alterations are driven by polyQ length-dependent metabolic alterations.
https://doi.org/10.1016/j.pneurobio.2023.102448

The MTM-HD study, a multi-omics analysis of HD patient peripheral tissues has been accepted by Genome Biology! We show abnormal molecular signatures of inflammation, energy metabolism and vesicle biology.
https://doi.org/10.1186/s13059-022-02752-5

TGFβ signaling in retinal neurons and Müller cells exhibits a neuroprotective effect and might pose promising therapeutic options to attenuate photoreceptor degeneration in humans.
https://doi.org/10.3390/ijms23052626

Read our pre-print about the MTM-HD study, a multi-omics analysis of HD patient peripheral tissues. We show abnormal molecular signatures of inflammation, energy metabolism and vesicle biology.
https://doi.org/10.1101/2022.02.03.475821

The integrity of the nucleolus is an important contributor to the formation of HTT aggregates. Monitoring this could prove to be a biomarker for HD progression.
https://doi.org/10.1038/s41419-021-04432-x
Read how targeting the Tau protein might have therapeutic potential in ALS (Amyotrophic Lateral Sclerosis)
https://doi.org/10.1007/s12035-021-02557-w

We have analysed the transcriptional changes in a model for retinits pigmentosa, a neurodegenerative disease of the retina.
https://doi.org/10.3390/ijms22126307

Dr. Neueder successfully finished his Habilitation in the field of experimental neurology at Ulm University.

Our paper about the role of SRSF6 in the splicing events of HTT just got published.
https://doi.org/10.1038/s41598-020-71111-w

Read our review about mitochondrial biology in HD.
https://doi.org/10.2217/nmt-2019-0033

Dr. Neueder successfully finished his BW-HDZ 'Baden-Württemberg-Zertifikat für Hochschuldidaktik'.
Read about it here.

Dr. Neueder became a member of the International Graduate School in Molecular Medicine Ulm (IGradU) junior faculty.
Read about it here.

Our paper showing that phenotype onset in Huntington’s disease knock-in mice is correlated with the incomplete splicing of the mutant huntingtin gene is published in the Journal of Neuroscience Research. Read it here: https://doi.org/10.1002/jnr.24493

A new review about RNA-Mediated Disease Mechanisms in Neurodegenerative Disorders is published and accessible from here:
https://doi.org/10.1016/j.jmb.2018.12.012

Our new paper about mechanisms that contribute to HTT mis-splicing got accepted by Nature Communications!
Accessible from here:
https://doi.org/10.1038/s41467-018-06281-3

Website is running
© 2024 All right reserved.